Introduction
In many experimental designs, model parameters vary across conditions or subjects and are conveniently stored in tabular (i.e., spreadsheet) form. In most cases, this kind of information is loaded into R as a data frame. This vignette demonstrates how to use a data frame to generate multiple predictions quickly.
We will walk through the full process using the package’s included
subopt_avian data set as an example.
The subopt_avian Data Set
Dunn et al. (2024) compiled the results of an extensive collection of studies investigating suboptimal choice behaviour. This dataset is included in the SiGN package.
The full data set is available as subopt_full. For model
evaluation purposes, however, Dunn et al. (2024) focused on a curated
subset of studies involving pigeons and starlings—particularly pigeons,
which were the most extensively studied and yielded the most robust,
consistently replicated findings. This filtered dataset is provided as
subopt_avian.
Each row in subopt_avian corresponds to a unique study
condition. Many of its columns—specifically columns 9 through 24—map
directly to the parameters expected by the SiGN() model
function (see the subopt_avian documentation for full
details on the column structure).
library(SiGN)
# Display column names
names(subopt_avian)
#> [1] "row" "study" "year" "species"
#> [5] "exp" "condition" "n" "cp"
#> [9] "il_dur_a" "il_dur_b" "tl_dur_a1" "tl_dur_a2"
#> [13] "tl_dur_b1" "tl_dur_b2" "tl_p_a1" "tl_p_a2"
#> [17] "tl_p_b1" "tl_p_b2" "tr_p_a1" "tr_p_a2"
#> [21] "tr_p_b1" "tr_p_b2" "il_sched_a" "il_sched_b"
#> [25] "tl_sched_a1" "tl_sched_a2" "tl_sched_b1" "tl_sched_b2"
#> [29] "forced_exposure" "DOI" "ref" "data_version"Of special note is column 8 ($cp), which contains the
observed choice proportion reported for each condition. These values can
be directly compared to the choice proportions predicted by the SiGN
model, which we will generate shortly.
Generating a List of Model Parameters
As detailed in the Get Started, to
generate model predictions with the SiGN() function, we
first need to prepare a list of model parameters in the format it
expects.
These parameters are currently stored as columns in the
subopt_avian data frame (specifically, columns 9 through
24). However, they are not yet in list form, and some required
inputs—such as s_delta, beta_log, and
beta_toggle—are not included in the data set. While these
defaults are not parameters most users need to adjust, they must be
present for SiGN() to run properly.
Rather than constructing this list manually, we use the
choice_params() function. This function not only formats
the input correctly but also performs important validation checks to
ensure that all required arguments are present and well-formed.
To convert the relevant columns into a list of named arguments, we
use do.call() in combination with as.list().
This allows us to pass the data frame columns directly into
choice_params() as if we had typed them out
individually.
# Store Model Parameters
#-------------------------------------------------------------------------------
# Extract relevant parameter columns (cols 9–24 align with choice_params())
data_cols <- subopt_avian[9:24]
# Construct model input list
params <- do.call(choice_params, as.list(data_cols))The do.call() function dynamically calls
choice_params() by unpacking the data frame columns into
named arguments. In other words,
do.call(choice_params, as.list(data_cols)) transforms each
column into an argument like il_dur_a = ...,
il_dur_b = ..., and so on—matching exactly what
choice_params() expects. This only works because the column
names in subopt_avian[9:24] are aligned with the argument
names that choice_params() requires.
Generating the Predictions
Now that we have constructed a valid list of model parameters, we can
pass it to the SiGN() function to generate predictions.
This function returns a variety of outputs, but here we are specifically
interested in the predicted choice proportions.
# Generate model predictions
#-------------------------------------------------------------------------------
preds <- SiGN(params)The SiGN model’s predicted choice proportion, for each row of
subopt_avian[9:24], can be called with
preds$cp. These values can be compared directly to the
observed choice proportions stored in subopt_avian$cp. For
more information about the SiGN() function and its outputs,
see the Get Started article.
Visualising the Predictions
To help visualise the model’s performance, we shall fit a simple linear regression to the observed versus predicted choice proportions. This provides a clear visual summary of their relationship and allows us to overlay both a fitted regression line and a 1:1 reference line. The closer the data points fall to the 1:1 line, the more closely the model’s predictions match the observed values—offering a quick and intuitive check on model accuracy.
# Fit a linear model comparing observed vs predicted
reg <- lm(subopt_avian$cp ~ preds$cp)
# Format annotation for plot
reg_txt <- sprintf(
"y = %.2f + %.2f x\n",
reg$coefficients[1], reg$coefficients[2]
)
library(ggplot2)
ggplot(mapping = aes(x = preds$cp, y = subopt_avian$cp)) +
# Add 1:1 reference line (dashed) to assess prediction accuracy
geom_abline(intercept = 0, slope = 1, linetype = 3) +
# Plot each observed vs. predicted data point as an open circle
geom_point(pch = 1, size = 1.75, stroke = 0.5) +
# Fit and overlay a linear regression line (no confidence band)
geom_smooth(method = "lm", se = FALSE) +
# Axis labels
labs(x = "Predicted", y = "Obtained") +
# Add regression summary text
annotate("text", x = 0.2, y = 0.9, label = reg_txt, size = 3.5) +
# Base theme
theme_bw(base_size = 12) +
# Make axis text black for visibility
theme(axis.text = element_text(colour = "black"))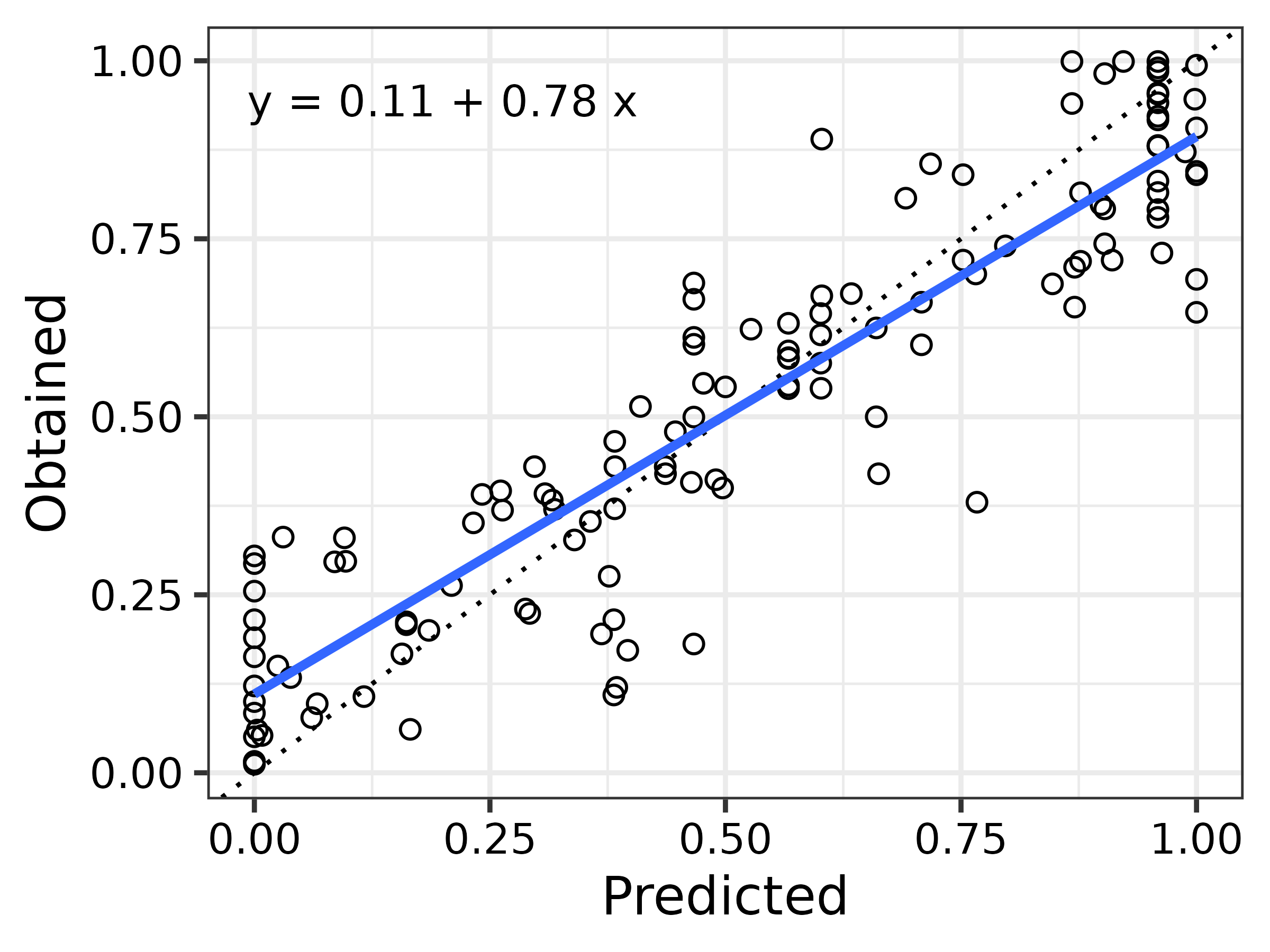
References
Dunn, R. M., Pisklak, J. M., McDevitt, M. A., & Spetch, M. L. (2024). Suboptimal choice: A review and quantification of the signal for good news (SiGN) model. Psychological Review. 131(1), 58-78. https://doi.org/10.1037/rev0000416